Many mechanistic aspects of LINE-1 and Alu retrotransposition are poorly understood. The Ghanim lab aims to understand how these two human mobile elements, LINE-1 and Alu, move through our genomes at a molecular level. The lab uses a combination of biochemical reconstitution, in vivo assays, and cryo-electron microscopy (cryo-EM) to address the important open questions in the field, outlined below.
An abundant mobile element in humans
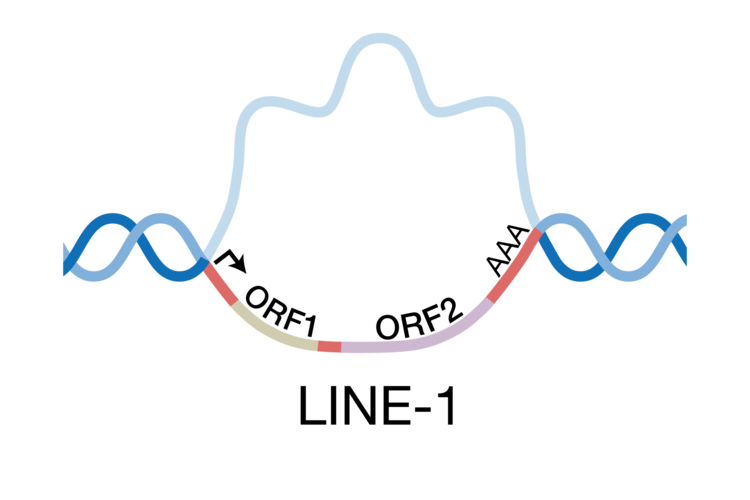
Nearly 30% of our genome is made up of two retrotransposable elements: long-interspersed nuclear element-1 (LINE-1) and the Alu element. Although most are inactive, a small subset of LINE-1s and Alus can still mobilize. This means that they can copy themselves from one location in and insert themselves into another. Consequently, LINE-1 and Alu mobility can be a significant mutagen. Their movement can result in numerous genetic diseases, is thought to drive oncogenic rearrangements in some cancers, and is linked to age-associated inflammation.
How does LINE-1 retrotranspose at the molecular level?
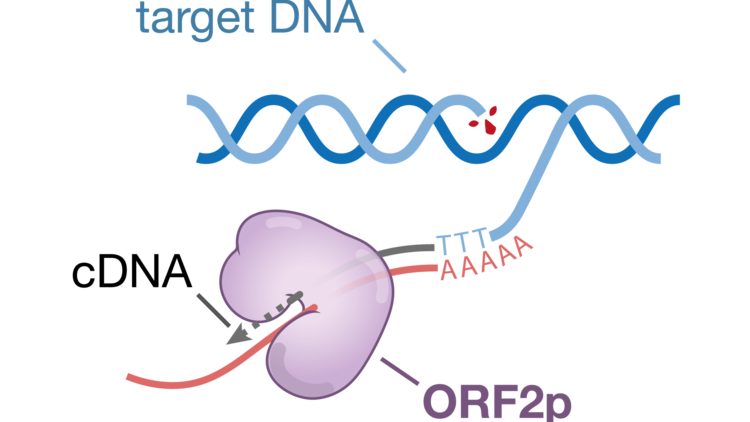
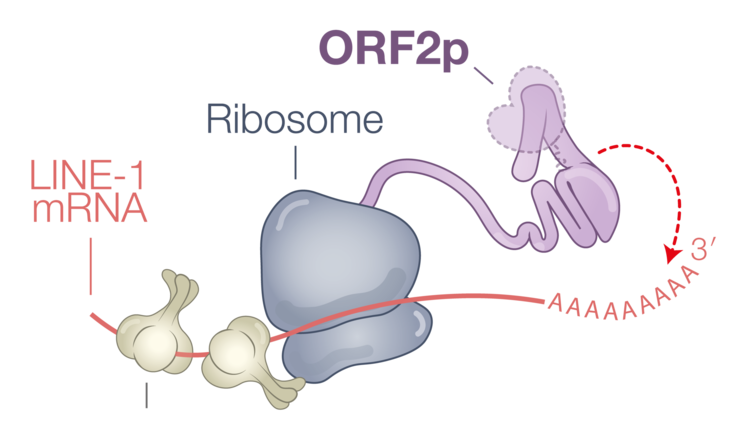
The LINE-1 mRNA encodes two proteins, ORF1p and ORF2p, that will assemble with the LINE-1 mRNA to form a retrotransposition competent LINE-1 ribonucleoprotein (RNP). During insertion, ORF2p nicks a target DNA, exposing a 3’ end which subsequently primes reverse transcription of the mRNA. Nicking of the second target DNA strand followed by second strand synthesis results in a new genomic insertion of the LINE-1 element.
The molecular mechanisms underlying LINE-1 retrotransposition remain largely unknown. For example, it is unclear how the LINE-1 RNP engages the target DNA for nicking by the endonuclease domain of ORF2p. It is also unknown how the nicked DNA strand is transferred to the reverse transcriptase domain for reverse transcription or how second strand synthesis is initiated.
How does the Alu element retrotranspose at the molecular level?
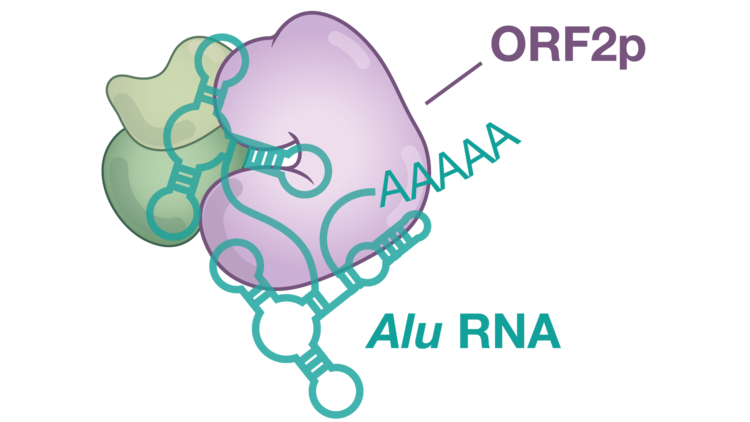
Alu elements do not encode retrotransposition factors; instead, they form structured RNPs that “capture” the LINE-1 ORF2 protein, enabling their own mobility. Again, numerous aspects of Alu element mobility are mechanistically unclear. For instance, the full composition of an Alu RNP is unclear, although it is expected to contain two proteins called SRP9 and SRP14, from the ribosomal signal recognition particle. It is also unclear how the Alu RNP captures ORF2p.
What is the molecular basis of cis-preference?
The LINE-1 machinery preferentially reverse transcribes its own encoding mRNA; not other LINE-1 mRNAs, not other cellular mRNAs. This characteristic is called cis-preference. This ensures the dissemination of retrotranspositionally competent elements and prevents the unproductive mobilization of other sequences, such as dysfunctional LINE-1 transcripts or the overwhelmingly abundant cellular RNAs. How this selectivity is established remains unclear, but evidence suggests that it requires a poly(A) tract and occurs co-translationally, coinciding with LINE-1 and Alu RNP assembly.
Can we turn these elements into tools?
There has been a resurgence in the development of mobile genetic elements into tools for biotechnology. Retrotransposons and their components hold great potential for applications in genome engineering but are underutilized compared to their DNA transposon counterparts. Mechanistic and structural insights will be necessary to engineer LINE-1 and Alu elements as molecular biology tools: i.e., modifying their substrate RNA or target DNA specificity.